DREAMS: deep read-level error model for sequencing data applied to low-frequency variant calling and circulating tumor DNA detection, Genome Biology
Descrição
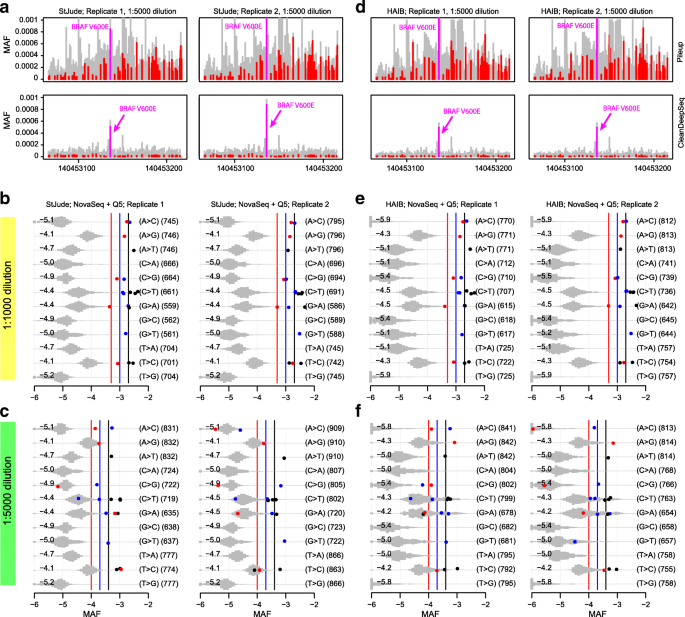
Circulating tumor DNA detection using next-generation sequencing (NGS) data of plasma DNA is promising for cancer identification and characterization. However, the tumor signal in the blood is often low and difficult to distinguish from errors. We present DREAMS (Deep Read-level Modelling of Sequencing-errors) for estimating error rates of individual read positions. Using DREAMS, we develop statistical methods for variant calling (DREAMS-vc) and cancer detection (DREAMS-cc). For evaluation, we generate deep targeted NGS data of matching tumor and plasma DNA from 85 colorectal cancer patients. The DREAMS approach performs better than state-of-the-art methods for variant calling and cancer detection.
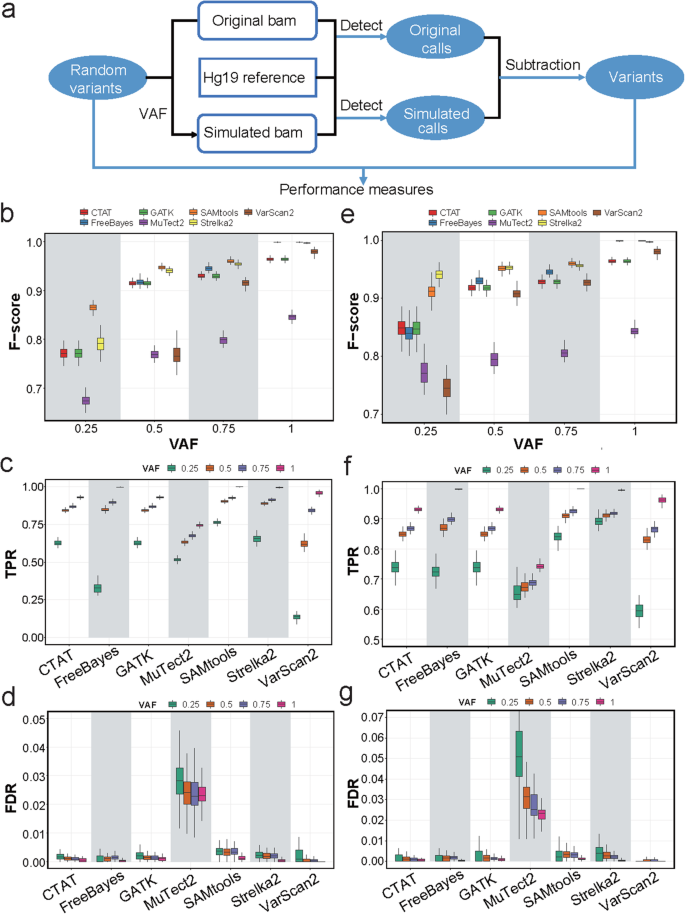
Systematic comparative analysis of single-nucleotide variant

Somatic small-variant calling methods in Illumina DRAGEN
PDF) DREAMS: deep read-level error model for sequencing data
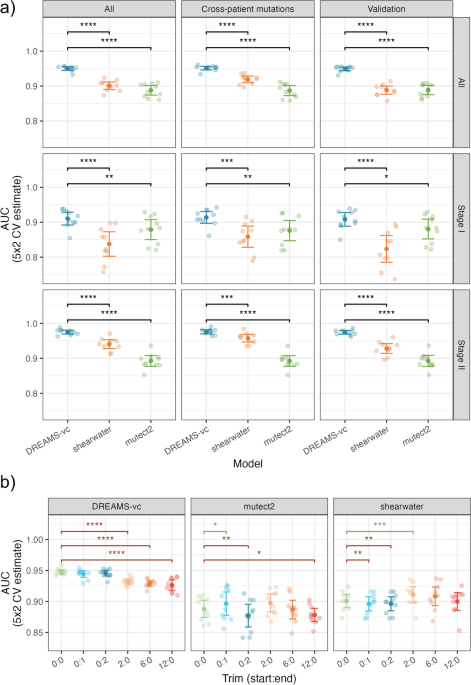
DREAMS: deep read-level error model for sequencing data applied to

DREAMS: Deep Read-level Error Model for Sequencing data applied to
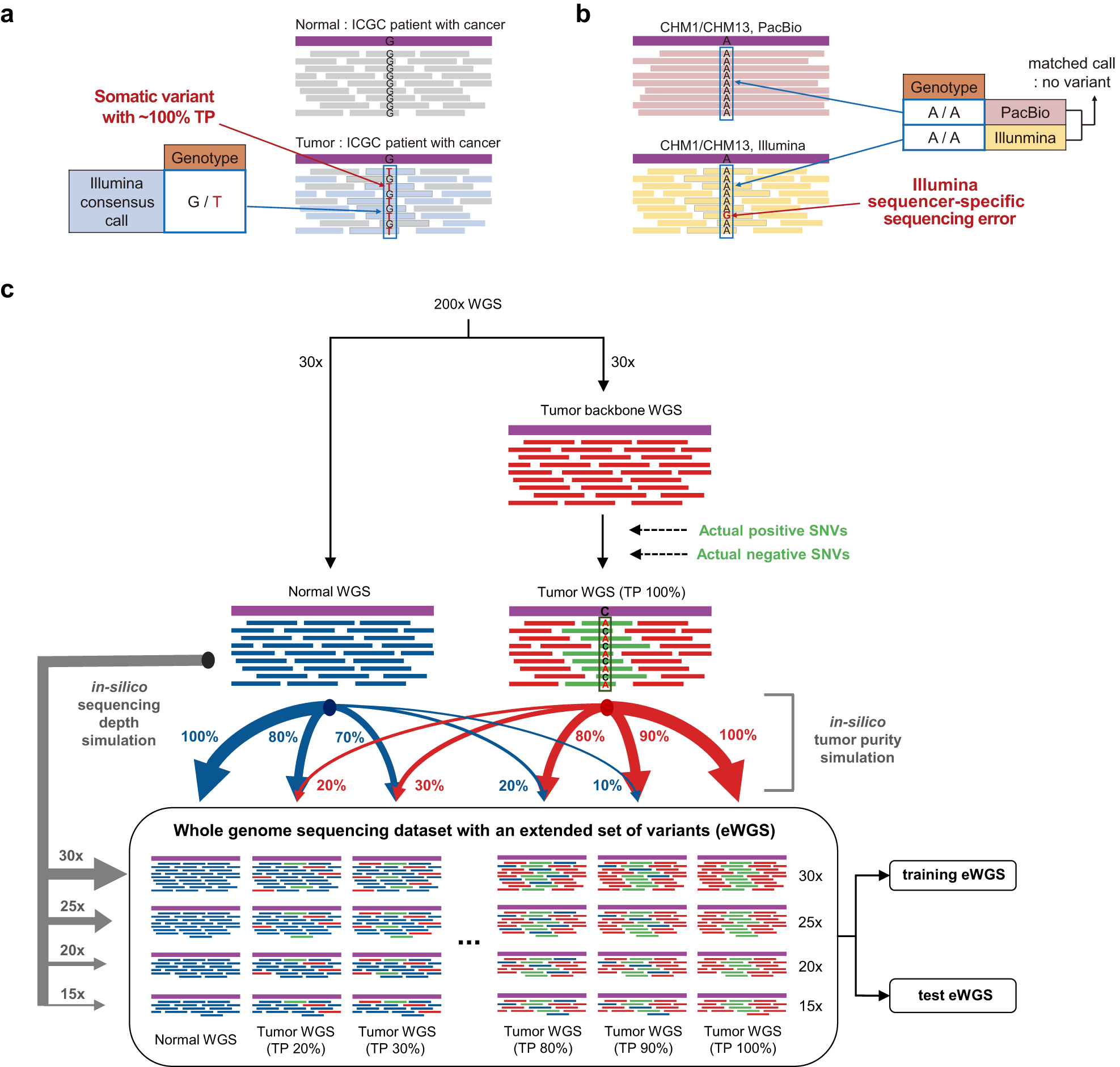
AIVariant: a deep learning-based somatic variant detector for

Machine learning guided signal enrichment for ultrasensitive
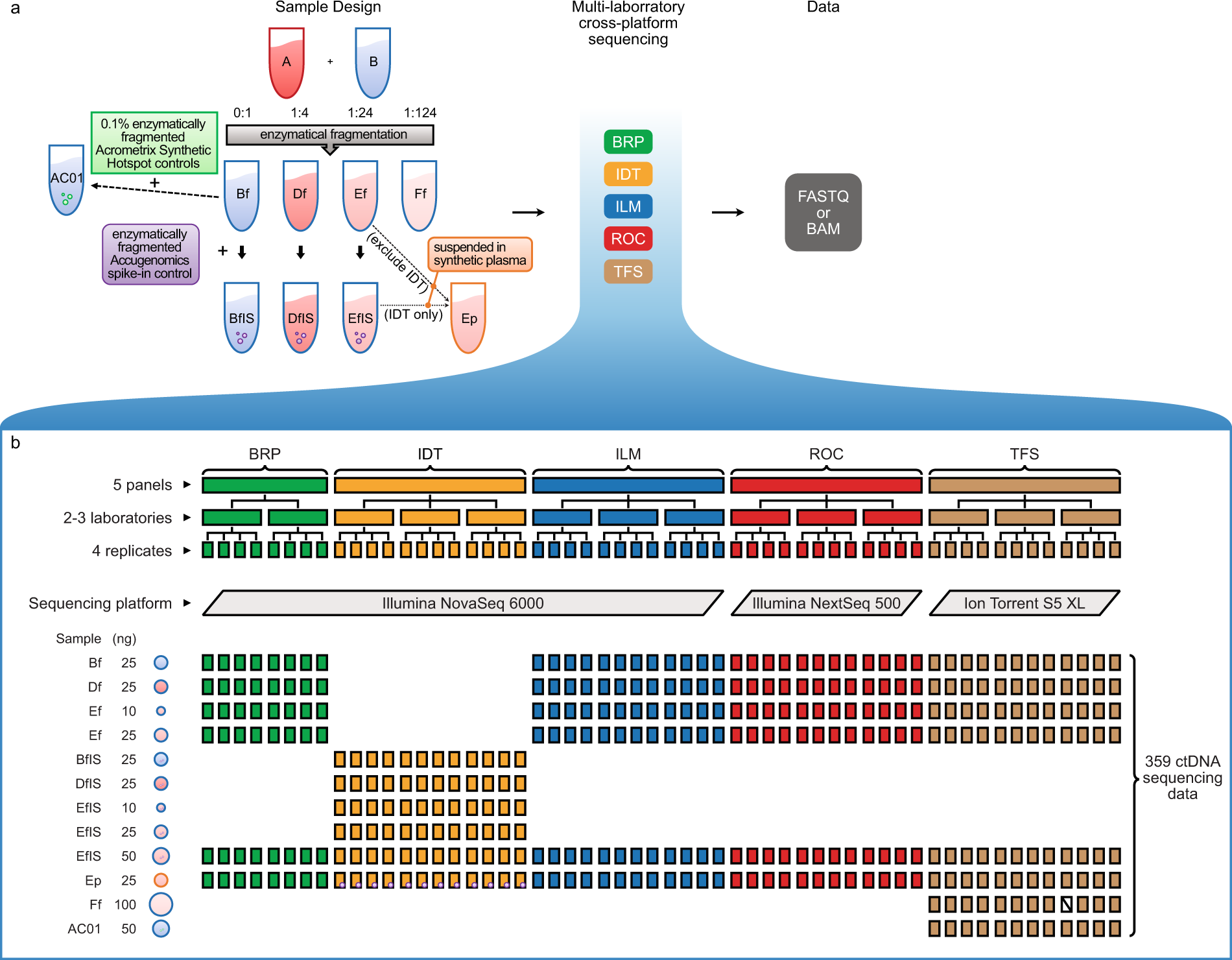
Ultra-deep sequencing data from a liquid biopsy proficiency study
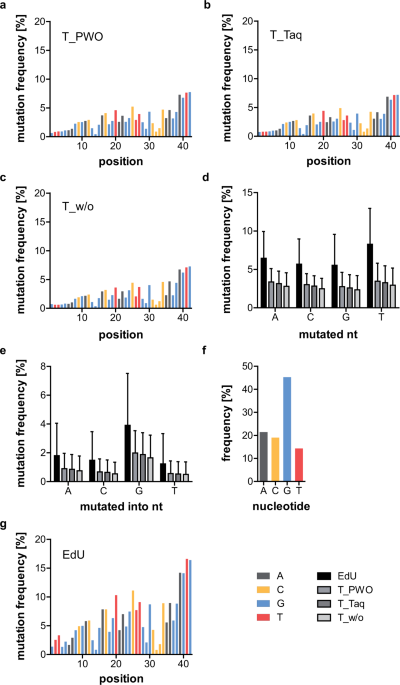
Systematic evaluation of error rates and causes in short samples
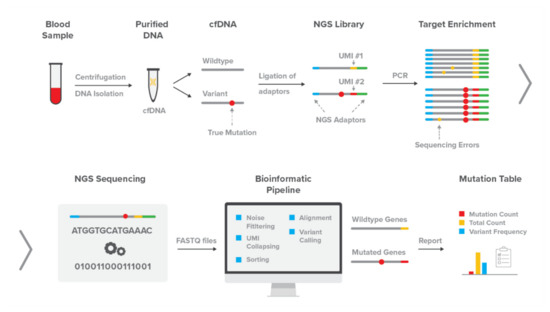
Genes, Free Full-Text
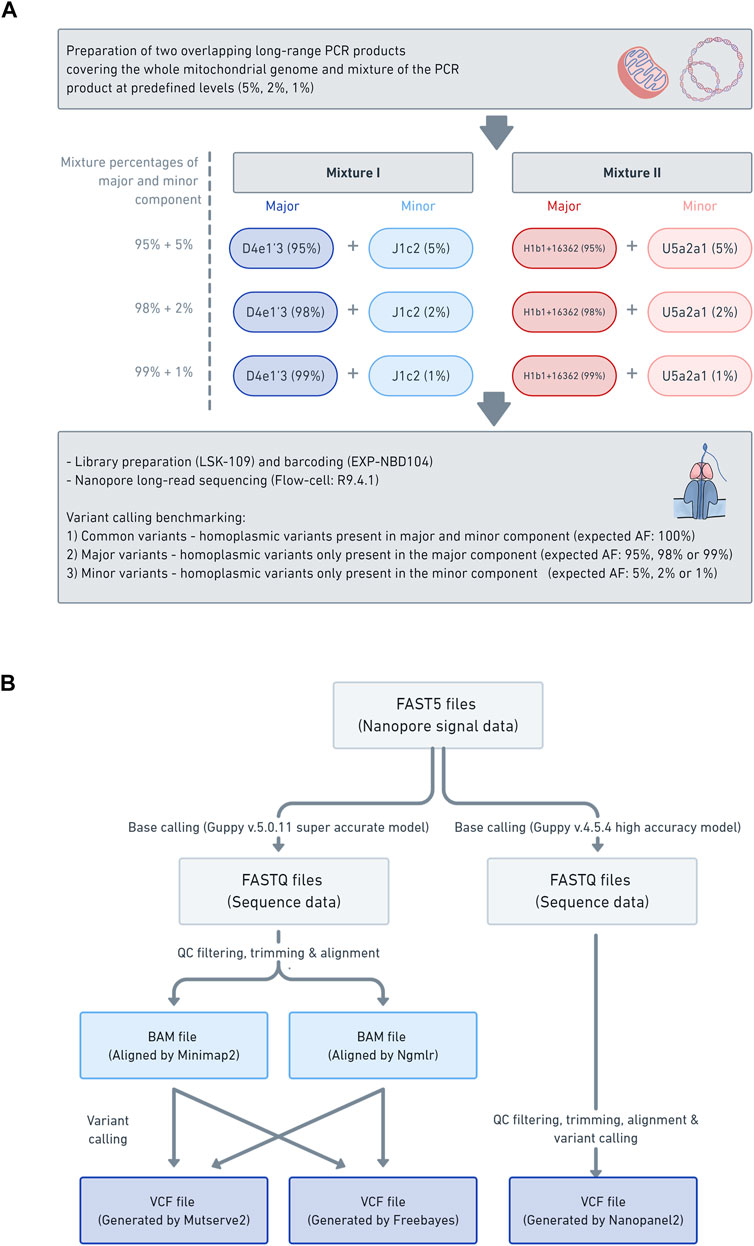
Frontiers Benchmarking Low-Frequency Variant Calling With Long

Types of errors. A screenshot from the IGV browser [21] showing

Integration of intra-sample contextual error modeling for improved
Analysis of error profiles in deep next-generation sequencing data
de
por adulto (o preço varia de acordo com o tamanho do grupo)






