Multi-Descriptor Read Across (MuDRA): A Simple and Transparent Approach for Developing Accurate Quantitative Structure–Activity Relationship Models
Descrição

Research Molecular Modeling Lab @ UNC
GitHub - MikolajMizera/pyMuDRA: Python implementation of Multi

AutoQSAR: an automated machine learning tool for best-practice

Chembench: A Publicly Accessible, Integrated Cheminformatics

PDF] Molecular fingerprint‐derived similarity measures for

PDF] Molecular fingerprint‐derived similarity measures for

Ranking of input variables.

In silico toxicology: From structure–activity relationships
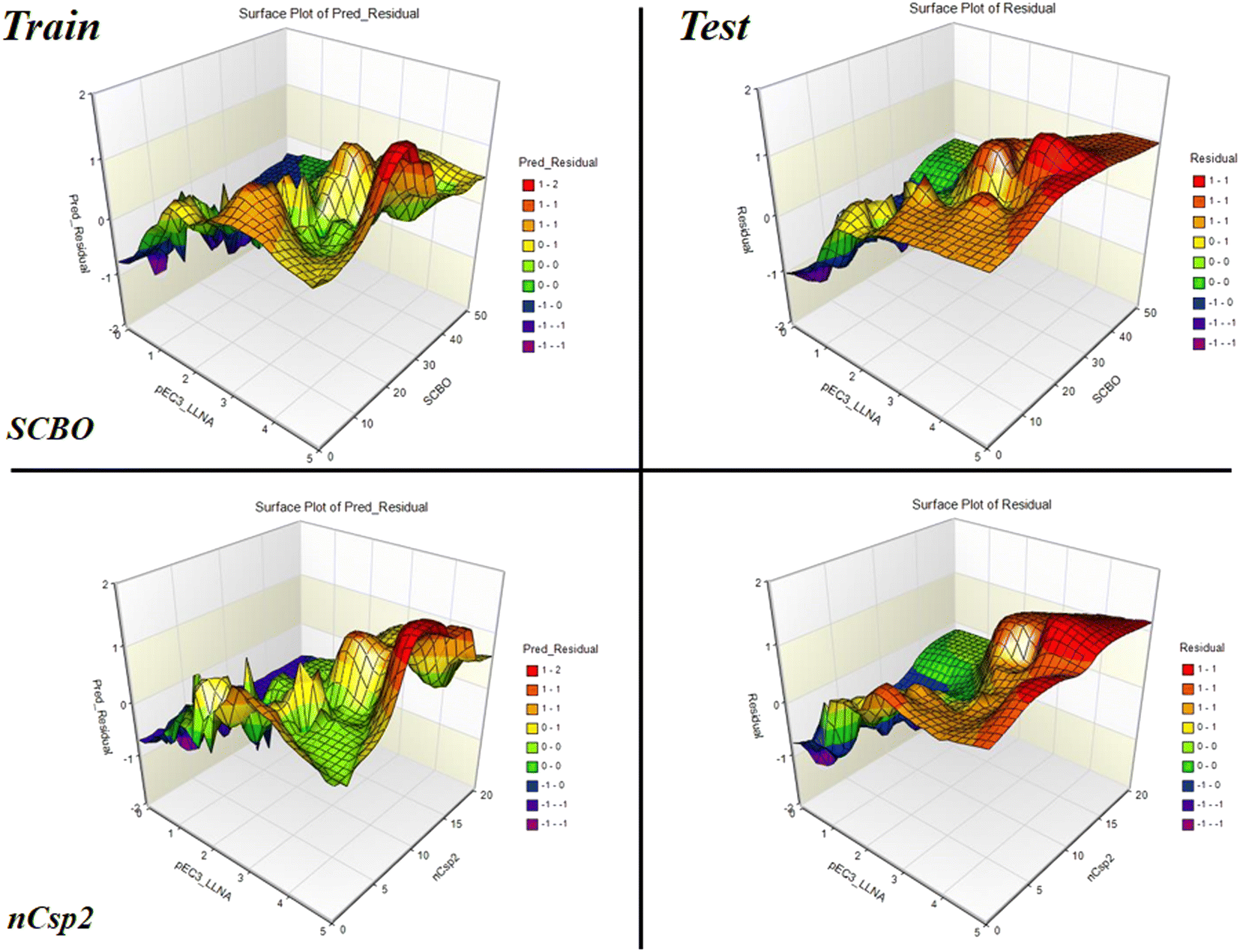
Read-across-based intelligent learning: development of a global q
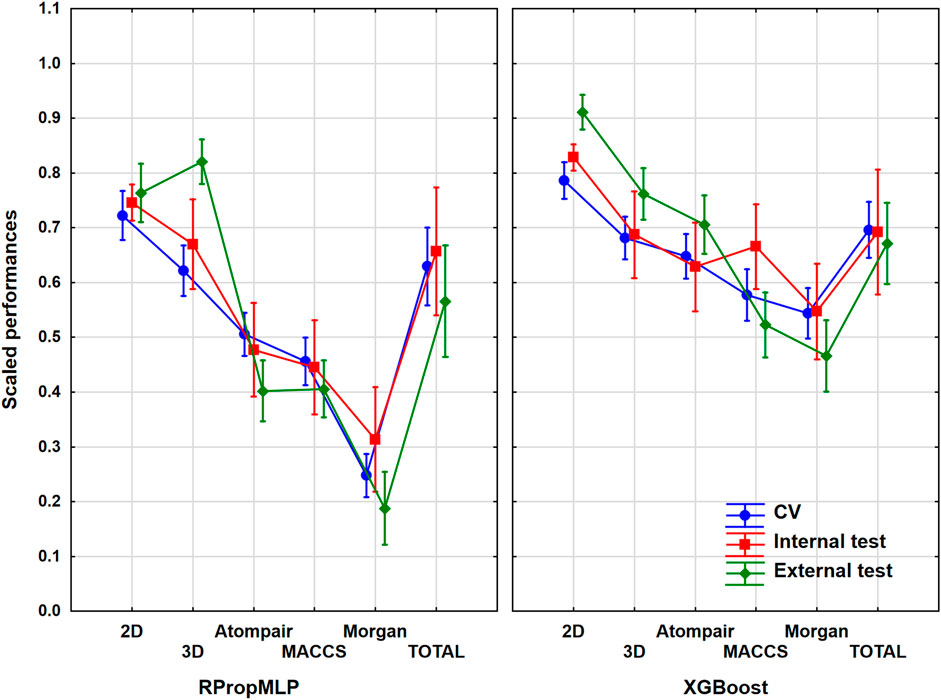
Frontiers Comparison of Descriptor- and Fingerprint Sets in

Read-across-based intelligent learning: development of a global q

ASAP] Novel Consensus Architecture To Improve Performance of Large

In silico toxicology: From structure–activity relationships
de
por adulto (o preço varia de acordo com o tamanho do grupo)







