PDF] Mining and Ranking Biomedical Synonym Candidates from
Descrição
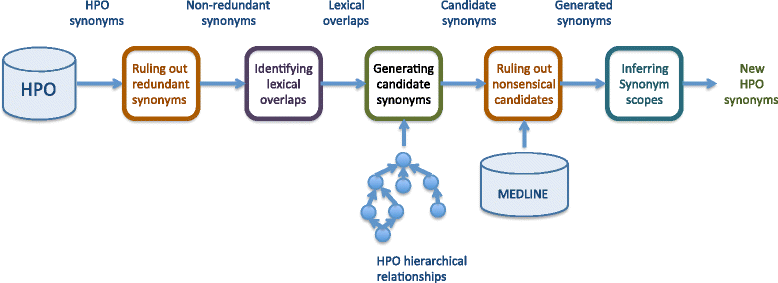
The results show that PRF-based reranking outperformed word embedding based approach and a strong baseline using interwiki link frequency, and suggests that medical synonyms mined from Wikipedia can increase the coverage of existing synonym resources such as UMLS. Biomedical synonyms are important resources for Natural Language Processing in Biomedical domain. Existing synonym resources (e.g., the UMLS) are not complete. Manual efforts for expanding and enriching these resources are prohibitively expensive. We therefore develop and evaluate approaches for automated synonym extraction from Wikipedia. Using the inter-wiki links, we extracted the candidate synonyms (anchor-text e.g., “increased thirst”) in a Wikipedia page and the title (e.g., “polyuria”) of its corresponding linked page. We rank synonym candidates with word embedding and pseudo-relevance feedback (PRF). Our results show that PRF-based reranking outperformed word embedding based approach and a strong baseline using interwiki link frequency. A hybrid method, Rank Score Combination, achieved the best results. Our analysis also suggests that medical synonyms mined from Wikipedia can increase the coverage of existing synonym resources such as UMLS.
![PDF] Mining and Ranking Biomedical Synonym Candidates from](https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41597-022-01920-3/MediaObjects/41597_2022_1920_Fig1_HTML.png)
A dataset for plain language adaptation of biomedical abstracts
![PDF] Mining and Ranking Biomedical Synonym Candidates from](https://ars.els-cdn.com/content/image/1-s2.0-S1570826822000403-ga1.jpg)
Comparison of biomedical relationship extraction methods and
![PDF] Mining and Ranking Biomedical Synonym Candidates from](https://www.researchgate.net/profile/Jinying-Chen/publication/301449060/figure/fig3/AS:361511215222796@1463202398536/Plot-of-Mean-Average-Precision-vs-N-for-relaxed-condition-N-is-the-number-of-queries_Q320.jpg)
PDF) Mining and Ranking Biomedical Synonym Candidates from Wikipedia
![PDF] Mining and Ranking Biomedical Synonym Candidates from](https://ars.els-cdn.com/content/image/1-s2.0-S153204642200257X-ga1.jpg)
An overview of biomedical entity linking throughout the years
![PDF] Mining and Ranking Biomedical Synonym Candidates from](https://www.mdpi.com/toxins/toxins-15-00192/article_deploy/html/images/toxins-15-00192-g001.png)
Toxins, Free Full-Text
![PDF] Mining and Ranking Biomedical Synonym Candidates from](https://www.researchgate.net/profile/Jinying-Chen/publication/301449060/figure/fig2/AS:361511215222794@1463202398330/Distribution-of-number-of-synonym-candidates-for-Wikipedia-Terms-N-is-the-number-of_Q320.jpg)
PDF) Mining and Ranking Biomedical Synonym Candidates from Wikipedia
![PDF] Mining and Ranking Biomedical Synonym Candidates from](https://ars.els-cdn.com/content/image/1-s2.0-S1532046422001150-ga1.jpg)
Graph-based abstractive biomedical text summarization - ScienceDirect
![PDF] Mining and Ranking Biomedical Synonym Candidates from](https://www.mdpi.com/energies/energies-16-00009/article_deploy/html/images/energies-16-00009-g001.png)
Energies, Free Full-Text
![PDF] Mining and Ranking Biomedical Synonym Candidates from](https://ars.els-cdn.com/content/image/1-s2.0-S1532046423000424-ga1.jpg)
Understanding common key indicators of successful and unsuccessful
de
por adulto (o preço varia de acordo com o tamanho do grupo)