Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
Descrição

Fast fit (A) and best fit (B) to the Catalyst common features model for

Development of an In Silico Prediction Model for P-glycoprotein Efflux Potential in Brain Capillary Endothelial Cells toward the Prediction of Brain Penetration
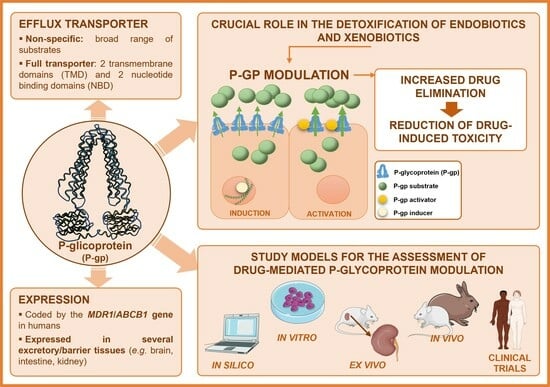
Molecules, Free Full-Text

Deep mutational scanning and machine learning reveal structural and molecular rules governing allosteric hotspots in homologous proteins
Plot of K p,brain and K p,uu,brain in the P-gp substrate before and
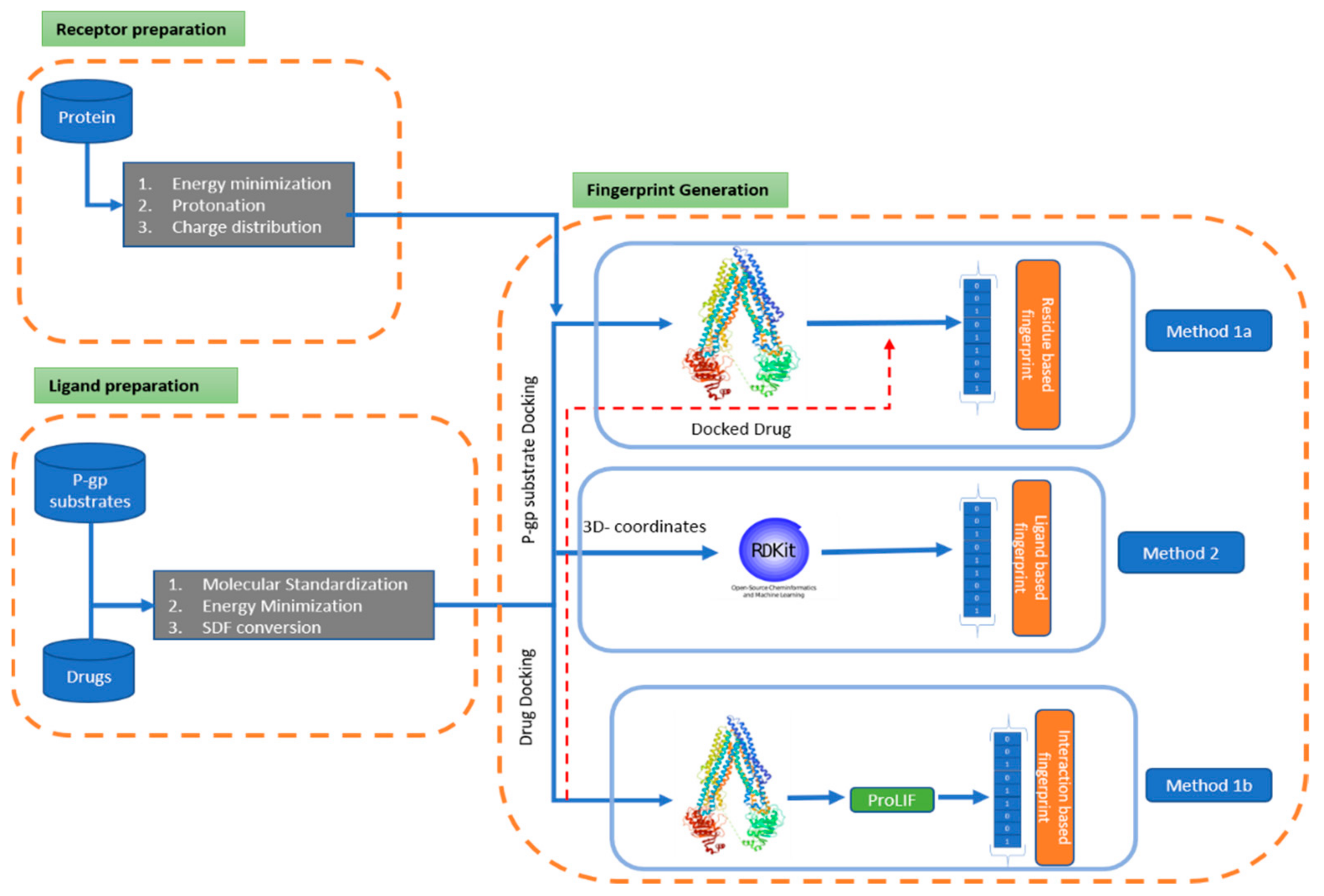
IJERPH, Free Full-Text
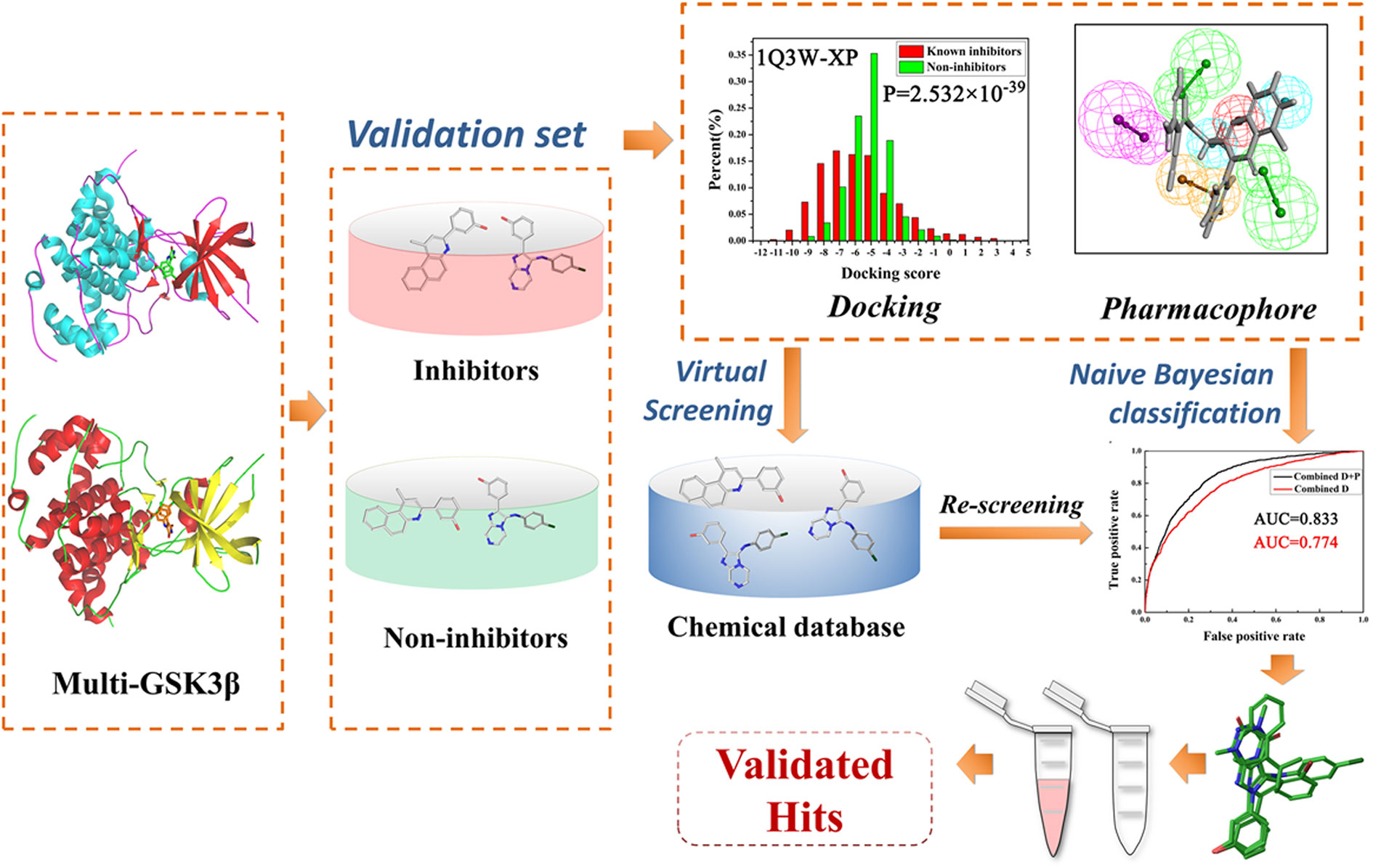
Frontiers Integrating Machine Learning-Based Virtual Screening With Multiple Protein Structures and Bio-Assay Evaluation for Discovery of Novel GSK3β Inhibitors
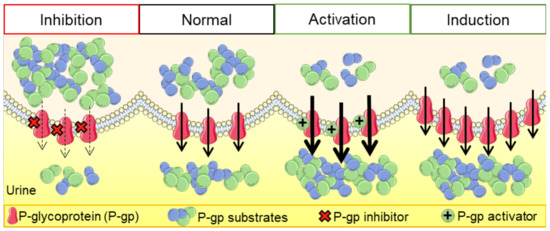
Molecules, Free Full-Text
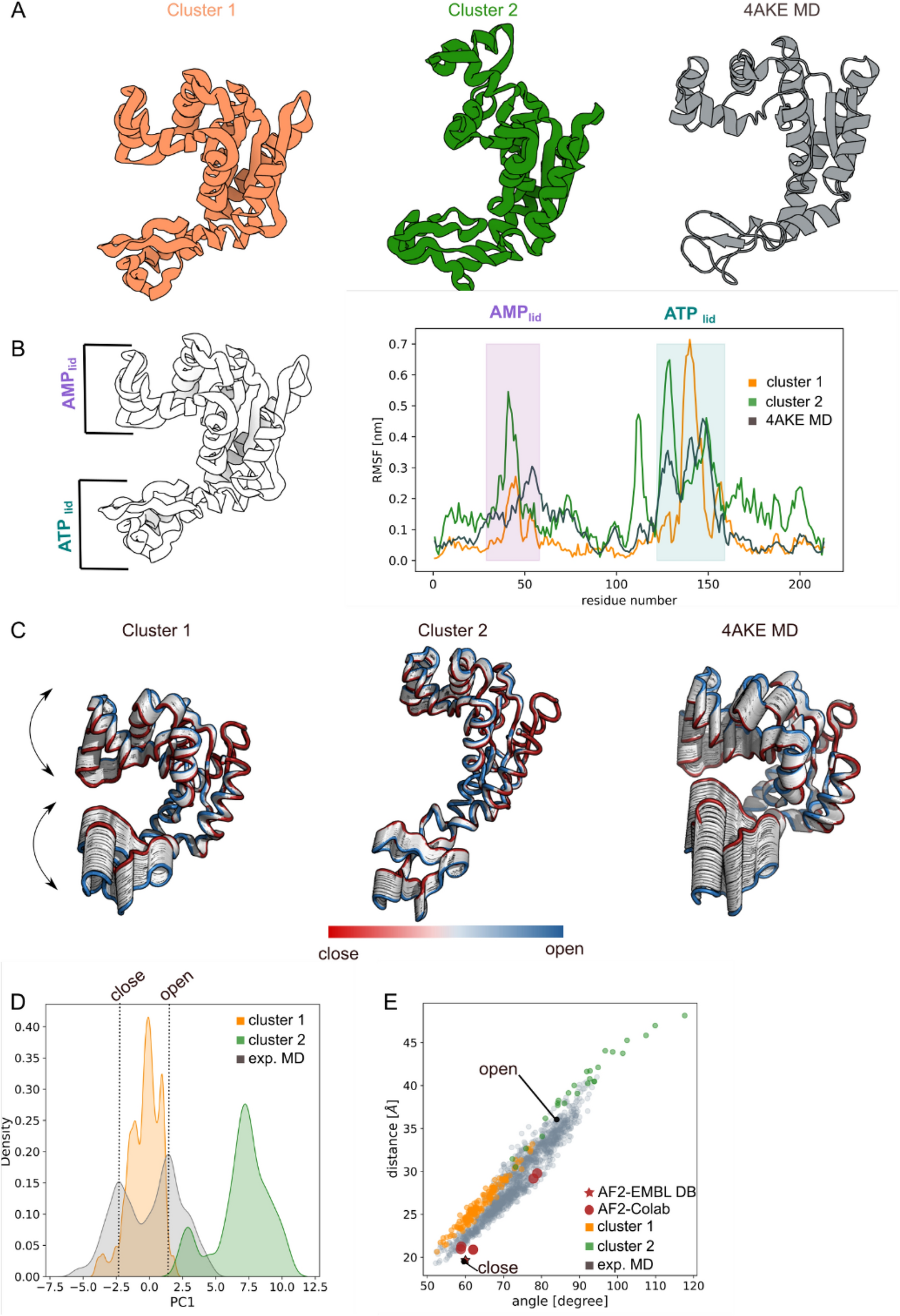
Machine learning/molecular dynamic protein structure prediction approach to investigate the protein conformational ensemble
de
por adulto (o preço varia de acordo com o tamanho do grupo)